
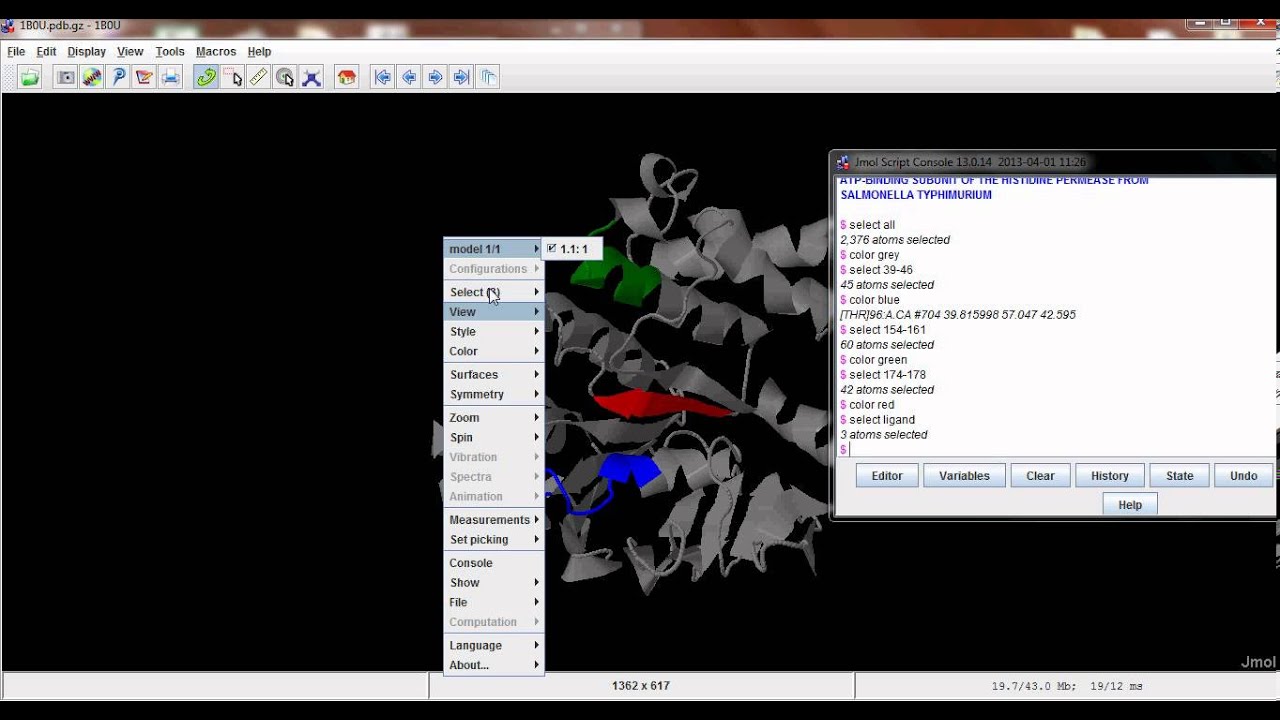
Radius of the spherical probe (in Angstroms) may be specified (default is 1.2). The solvent excluded surface ('isosurface solvent') is determined by rolling a probe solvent atom on the molecule at the distance of the van der Waals radii of the atoms.Use 'isosurface "filename.jvxl" ' to load the surface.ĭifferent types of surfaces may be specified (for more detail, see the Jmol Interactive Scripting Docs):.Copy the Jvxl file into the directory of the web page you are building so that the path to the Jvxl file can be easily linked.Use the 'write isosurface filename.jvxl' command to save the surface in a Jvxl file (this will save into the directory of your Jmol applet).Use one of the 'isosurface' commands (see below) to calculate the molecular surface of the selected molecule/s.Select the portion of your molecule you wish to display the surface of (e.g., "select nucleic" or "select ligand").Open the scripting window of the applet for command entry.
#JMOL SCRIPT FULL#
Versions 11.2 and up are recommended for full implementation of features below. To begin, open your Jmol Applet (Jmol.jar).One approach for doing this is outlined below, followed by a table with sample Jmol buttons and commands used to manipulate surface views. One can, however, use the Jmol applet to calculate a surface for a large molecule, save the isosurface calulations in a *.jvxl file ( JVoxel format), and quickly load the pre-calculated surface view (*.jvxl) using Jmol commands in a web page.
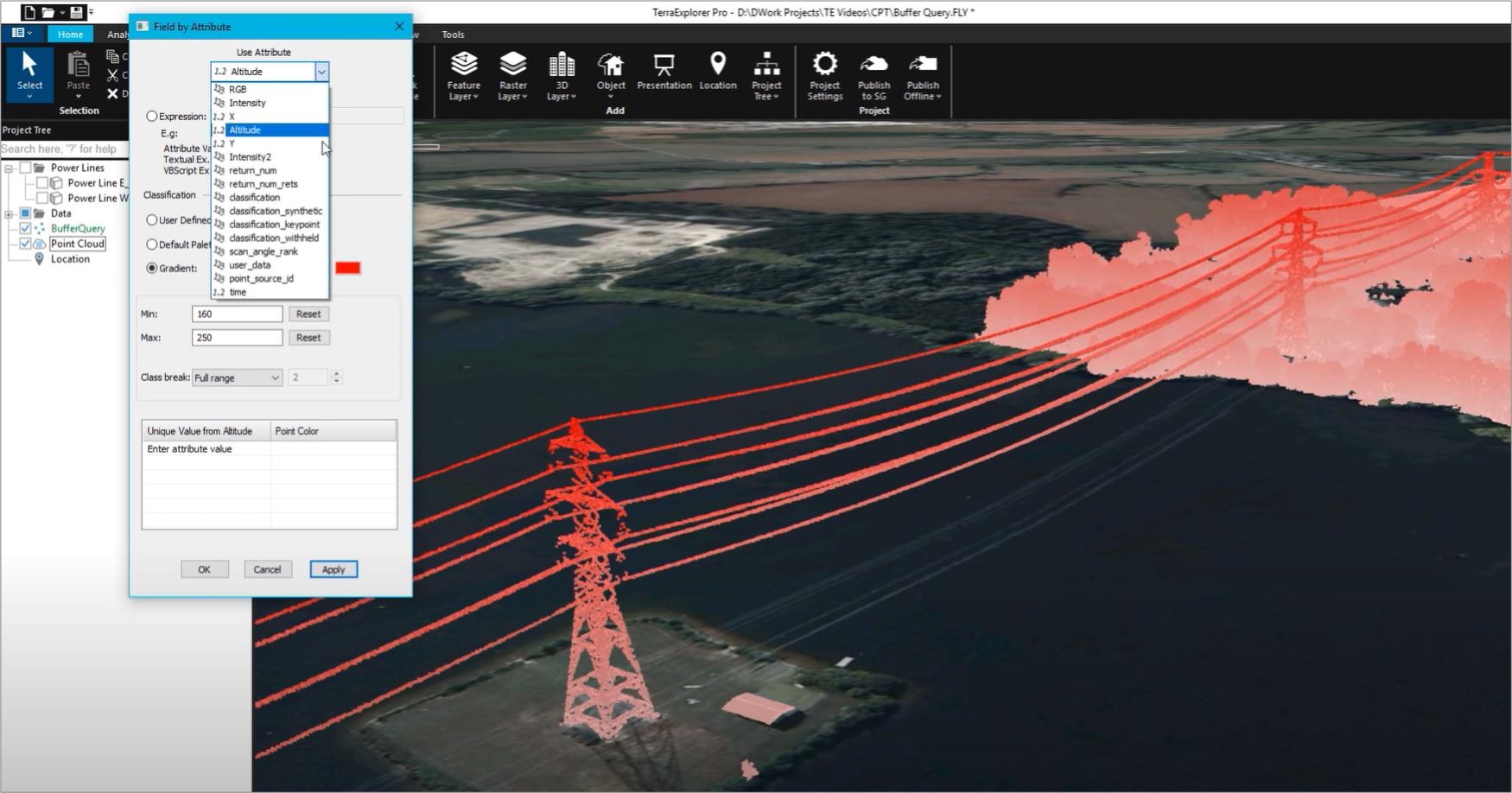
It is therefore not prudent to do this in web pages devoted to macromolecules, unless one is interested in displaying the surface of only a small subset of residues or atoms. Molecular surfaces can be calculated by Jmol, but for large molecules this can be quite CPU intensive and time consuming to do on the fly. you may need to do this several times, depending on the script.įor a more comprehensive treatment, see the Jmol If you get an unresponsive script, please choose continue. This is especially true now that we are using JSmol and HTML5 instead of the java-based Jmol. Note: surfaces require a lot of memory and are slow to load. This helps with funding opportunities for the OMM and helps ensure continued development of this resource. And please drop an email to with a link to your page/s.
Note: if this tutorial is helpful in the creation of your own webpages, please explicitly acknowledge your use of this page (URL, Author, date accessed, and a link to this page) in your webpage/s. Back to OMM Exhibits An Introduction to Jmol Surface Displays **


 0 kommentar(er)
0 kommentar(er)
